SOLUTION BRIEF
Big Memory Accelerates Single-Cell RNA Sequencing
Introduction
Modern gene sequencing technology can analyze millions of fragments of genetic material in parallel, thus generating results at a high throughput. An increasing focus is on understanding differential gene activity among individual cells using the techniques of single cell RNA sequencing (scRNA-seq).
A gene is a unique sequence of nucleotide base pairs that occur in a specific region of DNA. Not all genes are active at the same time. When a gene becomes active (i.e., expressed), its coding sequence is transcribed onto messenger RNA (mRNA). The complete collection of circulating mRNA molecules is referred to as the transcriptome. The more active a gene (higher expression), the more times that gene will be encountered during transcriptome sequencing.
After sequencing, the base pair sequence and number of observances of each gene per cell can be compiled. The assembled data forms the input to a series of computational steps in the analysis pipeline. Given the similarity of the pipeline steps to other machine learning problems, it is not surprising that the run times are similarly long and the pressures on memory and storage capacities similarly demanding.
The processes have long execution times and make stringent demands on the underlying compute resources, especially memory (DRAM) and storage. Volatile DRAM modules have limited capacity and require bulk loading of data before compute operations can begin. Persistent Memory (PMEM) is a recent technology breakthrough that enables high-capacity memory in a non-volatile DIMM. Memory Machine, a software package from MemVerge, can be used in conjunction with PMEM to provide a high-capacity memory pool with additional features that eliminate many bulk loads in the scRNA-seq analysis pipeline. The result is a dramatic decrease in execution times.
Memory Machine provides a memory virtualization and management layer that can be used to integrate traditional DRAM with Persistent Memory (PMEM), the latest development in non-volatile memory. The semiconductor technology underlying PMEM allows vendors to package PMEM in DDR4-compatible modules that have significantly higher capacity than DRAM without a proportionate cost increase. Although PMEM has natively higher latency compared to DRAM, Memory Machine is able to build a tiered memory hierarchy so that the average performance of DRAM is regained over the entire memory pool. Snapshots of the memory state can be taken without incurring any I/O or consuming additional memory, making the snapshot process highly efficient. The benefit of a memory snapshot is revealed when the time for writing intermediate stage results to durable storage and then reloading this data into memory at a subsequent stage can be avoided because the data is already resident in memory.
Single-Cell Sequencing: Tracking the Behavior of Individual Cells
High throughput sequencing is used increasingly to analyze gene expression in single cells, for example, to study rare cells, cells in a heterogeneous group, or even the variability in gene expression in a homogeneous group. Specifically, single-cell RNA sequencing (scRNA-seq)(i) can be used to track the trajectories of cells in development, i.e., the behavior of individual cells can be observed rather than an average across the entire population. While the exact data and techniques vary depending on the biological question of interest, most analyses follow a similar workflow.(ii)
The Raw Data
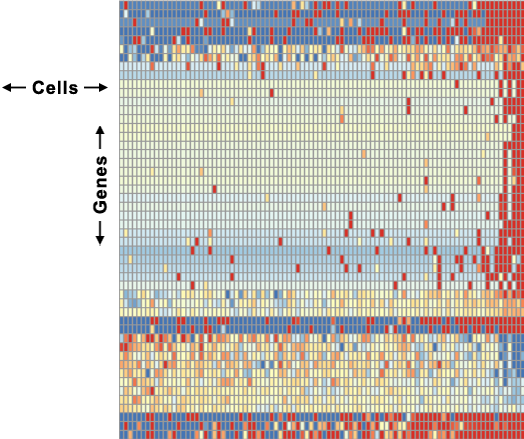
In scRNA-seq, the mRNA in single cells is fragmented and converted into a library of labeled mRNA segments that are then sequenced. The output is generally in the form of a two-dimensional matrix (the count matrix) in which the rows represent the genes that are of interest in the study, and the columns represent the individual cells in the sample. The matrix entries are the counts of the number of times each gene is identified in each cell, which is a relative measure of gene expression. A visual representation of a count matrix as a heatmap is shown in Figure 1.
Figure 1. Count matrix represented as a heatmap.
scRNA-sq Workflow
A database(iii) of the tools available for analyzing scRNA-sq data (currently over 800 tools) is publicly available. Over 80% of the tools use R or Python as the underlying platform. After the count matrix has been assembled, the subsequent workflow generally proceeds in the following steps.(iv)
1 – Quality Control (QC): scRNA-seq data are inherently noisy with anomalies introduced by the preparation of the samples and stochastic variation across cells. QC removes data from cells that may have been damaged or counts from incomplete sequencing reads.
2 – Normalization: Individual cell counts are normalized to eliminate cell-specific biases so that direct comparisons across cells can be made in later steps.
3 – Feature selection: The aim is to reduce the noise from uninteresting genes by only retaining genes that are likely to be responsible for heterogeneity across cells.
4 – Dimension reduction: The data is compacted and noise reduced further by transforming features into a smaller number of factors that still capture the majority of the data relationships. Principal components analysis (PCA) is typically used as the initial step followed by more aggressive pruning.
5 – Clustering: Cells are grouped according to similarities in their (normalized) gene expression counts. The groupings are used as proxies for distinguishing biological states.
The specific number of tasks and division into steps depends on the scRNA-seq analysis tools used: some tools perform only a single step in the process; other tools perform multiple steps. Depending on the toolkit used, the workflow described above may be further sub-divided (for example, in the study described below 11 steps are used).
When Data is Greater Than Memory
As single cell analysis techniques improve, more cells can be included in a single experimental run with a resultant increase in the size of the count matrix. As is customary for machine learning, several steps in the analysis are iterative in order to tune model parameters. Iteration requires that data saved from a previous step be reloaded into memory to become input for the next cycle. Furthermore, computations involving the count matrix or derivatives require that the data reside in memory. If the memory capacity is insufficient, then the computations must proceed in batches. Both memory capacity and bandwidth are key factors in determining the time for a complete scRNA-seq analysis.
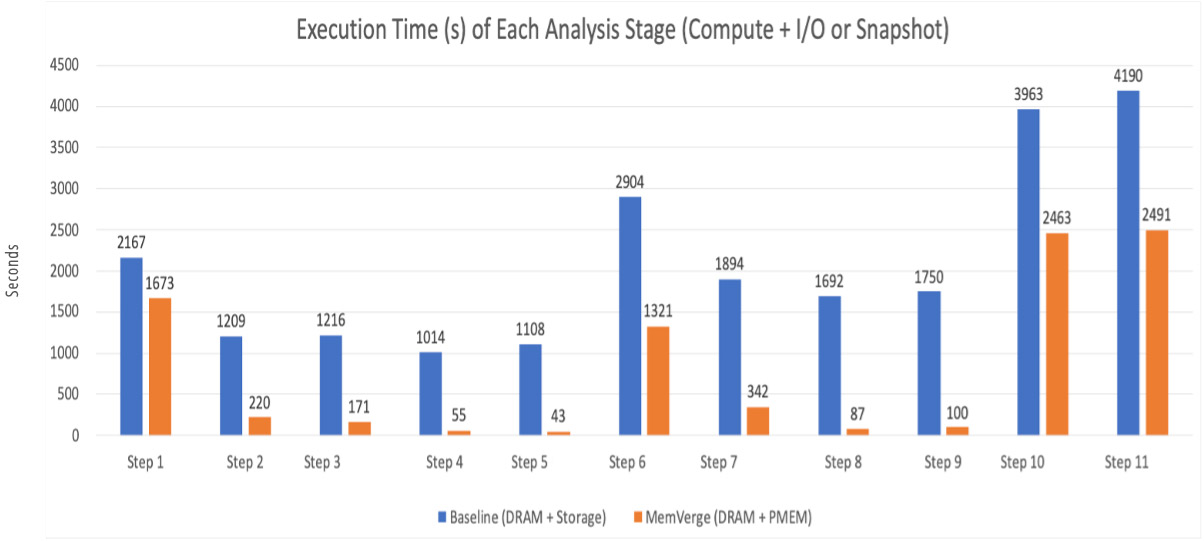
1,000x scRNA-sq Load Times, 25x Faster Execution
To investigate the impact of Memory Machine on the performance of an scRNA-seq analysis (Mouse Cell Atlas, count matrix size 31787 x 813348), a high end server (2 x Intel Gold 18 core CPUs @2.60GHz) running Centos was loaded with 12 x 16 GB modules of DDR4 memory and 12 x 128 GB modules of Intel Optane DC PMEM. As a baseline, the complete analysis using R-based tools was completed using DRAM only. The analysis was repeated using Memory Machine and the combined DRAM plus PMEM memory pool.
The results show significant performance improvements when using the DRAM plus PMEM combination. For each step, data load times decreased from ~1000s to ~1s by using memory snapshots. Figure 2 shows up to 25x improvement in the time to reload data into memory in the iterative steps.
Figure 2. Improvement in Execution Time
Real-Time Sequencing Can Help Control Pandemics
Since DRAM was invented in 1969, the server memory model has changed little as DRAM continues to be expensive, volatile, and with higher capacities achieved only by constant IO to slower storage used as an extension of memory.
With Big Memory, including Intel Optane Persistent Memory plus Memory Machine software, scientists can now track the dispersal of a virus in the present, gene sequencing can reconstruct the processes that drove its global spread in the past and determine when it first arose within a population.
Single-cell RNA sequencing analysis is a pipeline that includes tasks typically found in machine learning. The process imposes significant demands on memory resources (computation uses very large matrices that need to fit in memory), on storage (intermediate results must be saved and reloaded for other stages), and the process is long running (compute intensive and many stages are repeated for parameter tuning). By using Memory Machine and its snapshot capability the overall time to execute the entire scRNA-seq analysis can be reduced significantly. By extension, other bioinformatics analyses that use large matrices derived from next-generation sequencing techniques can also be accelerated.
(i) Luecken, MD and Theis, FJ. Current best practices in single-cell RNA-seq analysis: a tutorial, Mol Syst Biol 2019 (doi: https://www.embopress.org/doi/full/10.15252/msb.20188746)
(ii) https://pubmed.ncbi.nlm.nih.gov/32221477/
(iii) https://www.scrna-tools.org
(iv) https://github.com/Bioconductor/OrchestratingSingleCellAnalysis